rOpenSciコミュニティの紹介
2023-06-14

ニッタ ジョエル Ph.D.(生物学)
千葉大学 国際学術研究院 国際教養学部
https://www.joelnitta.com
今日話すこと
rOpenSciの紹介
rOpenSciに投稿した二つのパッケージの話
- dwctaxon https://github.com/ropensci/dwctaxon
- canaper https://github.com/ropensci/canaper
(統計パッケージ)
なぜrOpenSciに参加する(パッケージを投稿する)
と良いのか?
rOpenSciとは?
We help develop R packages for the sciences via community driven learning, review and maintenance of contributed software in the R ecosystem
![]()
Rパッケージを査読して、支援するコミュニティー
rOpenSciの範囲
R packages for the sciences
科学的な解析に使われるパッケージ
・・と言っても、結構広い
rOpenSciの範囲
- Computing infrastructure
(インフラ) - Databases
(データベース) - Geospatial(地理空間)
- Images(画像)
- Literature(文献)
- Security(セキュリティ)
- Statistics(統計)
- Taxonomy(分類)
rOpenSciのコミュニティー
2011年から
Staff(給料あり)6人
あとはボランティア

rOpenSciのスタッフ

Community Calls
Blog
なぜ投稿する(査読してもらう)?
自分のパッケージをよりよくする
自分のコードをよりよくする
JOSS(とMethods Ecol. Evol.)のコードレビューの代わりになる
rOpenSciからの援助をもらう(PR、コードに困ったとき)
管理が出来なくなったら、他のメンテナーを探してくれる
投稿について
- 重要文献:「rOpenSci Packages: Development, Maintenance, and Peer Review」
- 完全ガイドブック
- 投稿する前にGuide for Authorsを読むべし
- 査読のプロセスは全てオープンでGitHub上で行われる

投稿する前に
- もし自分のパッケージがrOpenSciの範囲(scope)にフィットするかどうか不安だったら、まずはpre-submission inquiryを出す(issueを開く)
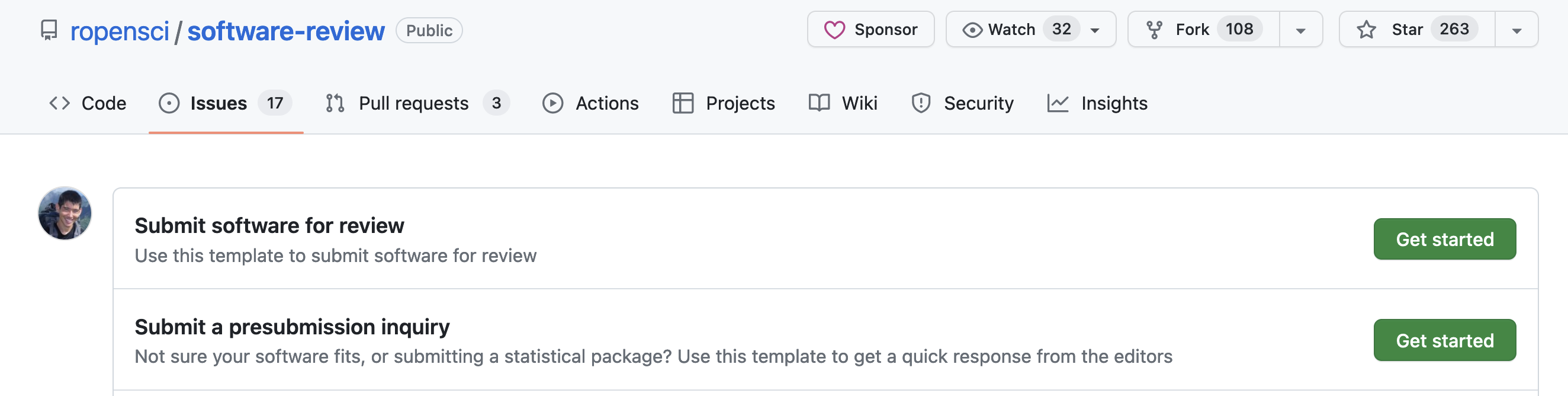
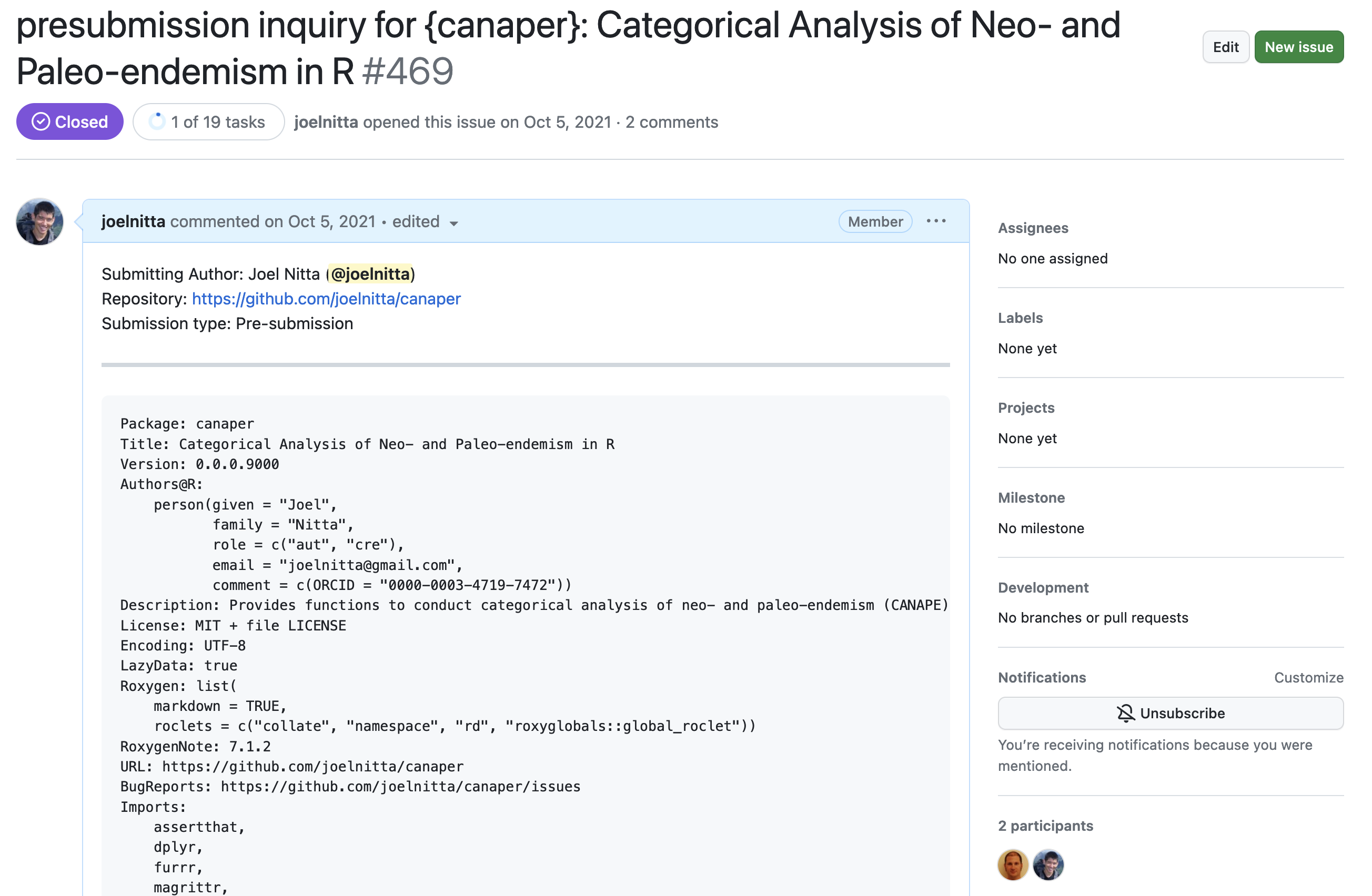
Pre-submission inquiryの例:canaper
投稿のプロセス:始まり
投稿のプロセス:始まり
DESCRIPTIONなどをhttps://github.com/ropensci/software-review/issues/に
ポストする基本的なチェックが自動的に行われる 🤖
投稿のプロセス:査読
エディターが二人のレビュアーを誘って、見てもらう(2週間)
コメントに答える(2週間)
レビジョンが認められたら、受かる 🎉
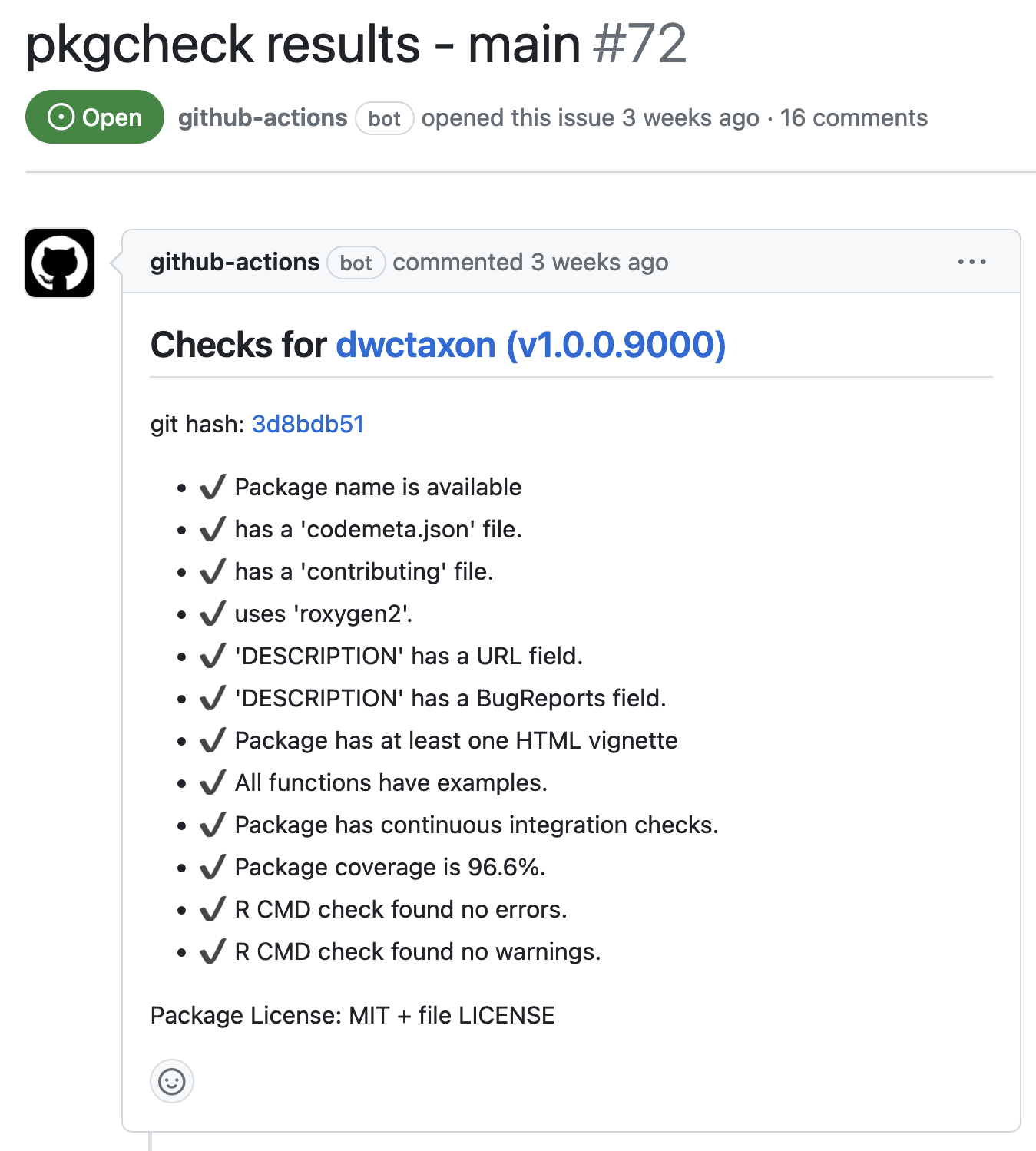
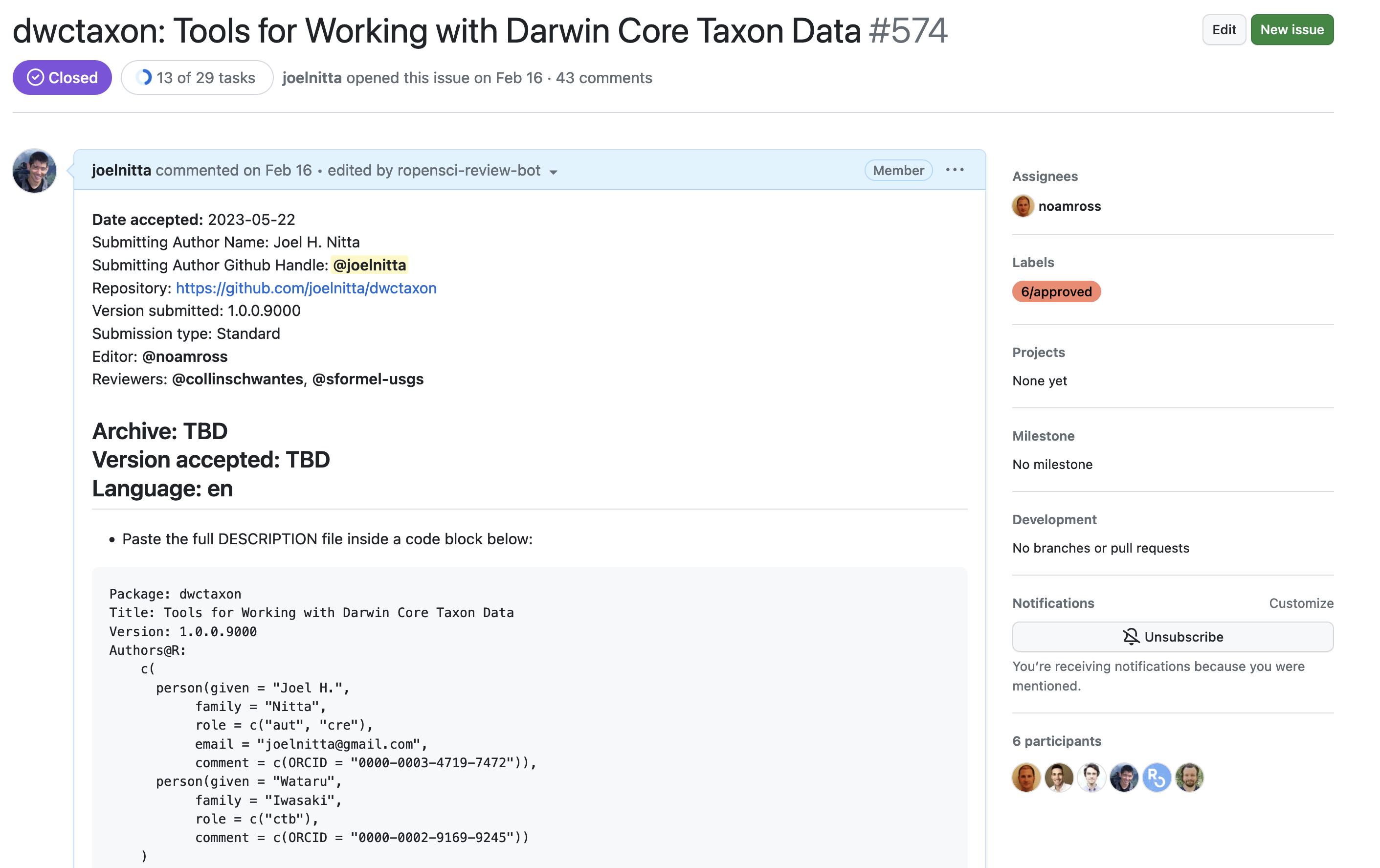
投稿の例:dwctaxon
投稿のプロセス:受かってから
https://github.com/ropensciにレポジトリーを移動する
パッケージのウエブサイトをhttps://docs.ropensci.org/に移動する
パッケージが自動的にr-universeに載る
CRAN・Bioconductorに載せる(任意)
JOSSに投稿する(任意)
統計的なパッケージ
統計解析を行なっているパッケージ
より細かい査読が行われる
統計的なパッケージの範囲
- Bayesian and Monte Carlo Routines
- Regression and Supervised Learning
- Dimensionality Reduction, Clustering, and Unsupervised Learning
- Exploratory Data Analysis (EDA) and Summary Statistics
- Time Series Analyses
- Machine Learning
- Spatial Analyses
統計的なパッケージのスタンダード
Standards are good
Standards should be strict
No-one reads standards
スタンダードは良い物である
スタンダードは厳しくすべし
誰もスタンダードなんて読まない
Colin Gillespie, European R Users Meeting 2020にて
統計的なパッケージのスタンダード
- 分野ごとに決まっている
例えば、Machine Learning:
ML1.0 Documentation should make a clear conceptual distinction between training and test data (even where such may ultimately be confounded as described above.)
https://stats-devguide.ropensci.org/standards.html#input-data-specification
srrパッケージでスタンダードを
管理する

srrパッケージでスタンダードを
管理する
- roxygen2のコメント
#'にタグをつける@srrstatsで始める
#' @srrstats {G2.1, G2.6} Check input types and lengths
assertthat::assert_that(
inherits(comm, "data.frame") | inherits(comm, "matrix"),
msg = "'comm' must be of class 'data.frame' or 'matrix'"
)srrパッケージでスタンダードを
管理する
devtools::document()する度にスタンダードが
チェックされる
> document()
ℹ Updating canaper documentation
ℹ Loading canaper
────────────────────────────────────── rOpenSci Statistical Software Standards ─────────────────────────────────────
── @srrstats standards (179 / 231):
* [G2.0a, G2.1a, G2.3b, G1.4, G1.4a] in function 'calc_biodiv_random()' on line#40 of file [R/calc_biodiv_random.R]
* [G1.3, G1.0, G1.4, G2.1, G2.6, G3.0] in function 'cpr_classify_endem()' on line#41 of file [R/cpr_classify_endem.R]
* [G1.3, G2.0a, G2.1a, G2.3b, G1.4, G2.1, G2.6, G2.0, G2.2, G2.3, G2.3a, G3.0] in function 'cpr_classify_signif()' on line#51 of file [R/cpr_classify_signif.R]
* [G2.0a, G2.1a, G2.3b, G1.4, G1.4a, G2.1, G2.6] in function 'cpr_iter_sim()' on line#65 of file [R/cpr_iter_sim.R]
* [G2.1, G2.6] in function 'cpr_rand_comm()' on line#54 of file [R/cpr_rand_comm.R]
* [G2.0a, G2.1a, G2.3b, G2.7, UL1.0, UL4.3a, G1.3, UL3.4, G1.0, G1.4, G2.0, G2.2, G2.1, G2.3, G2.3a, G2.4a, G2.6, G2.13, G2.14, G2.14a, G2.15, G2.16, UL1.1, G2.8, G2.8, UL1.2, UL1.2, G2.15, UL1.1, G2.11, UL1.1, G2.16, UL1.1, G2.4a, UL1.4, UL1.4, UL1.4, UL2.0, UL1.4, G2.1] in function 'cpr_rand_test()' on line#150 of file [R/cpr_rand_test.R]
* [G1.4, G5.1] in function 'acacia()' on line#28 of file [R/data.R]
* [G1.4, G5.1] in function 'biod_example()' on line#57 of file [R/data.R]
* [G1.4, G5.1] in function 'phylocom()' on line#87 of file [R/data.R]
* [G1.4, G5.1] in function 'biod_results()' on line#125 of file [R/data.R]
* [G1.4] in function 'mishler_signif_cols()' on line#145 of file [R/data.R]
* [G1.4] in function 'cpr_signif_cols()' on line#159 of file [R/data.R]
* [G1.4] in function 'cpr_signif_cols_2()' on line#174 of file [R/data.R]
* [G1.4] in function 'mishler_endem_cols()' on line#201 of file [R/data.R]
* [G1.4] in function 'cpr_endem_cols()' on line#223 of file [R/data.R]
* [G1.4] in function 'cpr_endem_cols_2()' on line#245 of file [R/data.R]
* [G1.4] in function 'cpr_endem_cols_3()' on line#267 of file [R/data.R]
* [G1.4] in function 'cpr_endem_cols_4()' on line#289 of file [R/data.R]
* [G2.0a, G2.1a, G2.3b, G1.4, G1.4a, G2.1, G2.6, G2.3, G2.3a] in function 'get_ses()' on line#43 of file [R/get_ses.R]
* [G1.2, G5.1, G5.7, UL7.1] on line#188 of file [R/srr-stats-standards.R]
* [G1.4, G1.4a, G2.1, G2.6] in function 'count_higher()' on line#18 of file [R/utils.R]
* [G1.4, G1.4a, G2.1, G2.6] in function 'count_lower()' on line#58 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function 'lesser_than_single()' on line#91 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function '%lesser%()' on line#111 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function 'lesser_than_or_equal_single()' on line#128 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function '%<=%()' on line#146 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function 'greater_than_single()' on line#163 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function '%greater%()' on line#183 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function 'greater_than_or_equal_single()' on line#200 of file [R/utils.R]
* [G3.0, G2.1, G2.6] in function '%>=%()' on line#218 of file [R/utils.R]
* [G5.3] on line#80 of file [tests/testthat/test-calc_biodiv_random.R]
* [G5.2, G5.2a, G5.2b, UL7.0] on line#23 of file [tests/testthat/test-cpr_classify_endem.R]
* [G5.4a, G5.5] on line#61 of file [tests/testthat/test-cpr_classify_endem.R]
* [G5.2, G5.2a, G5.2b, UL7.0] on line#3 of file [tests/testthat/test-cpr_classify_signif.R]
* [G5.4a, G5.5] on line#35 of file [tests/testthat/test-cpr_classify_signif.R]
* [G5.2, G5.2a, G5.2b, UL7.0] on line#3 of file [tests/testthat/test-cpr_iter_sim.R]
* [G5.4, G5.5] on line#35 of file [tests/testthat/test-cpr_iter_sim.R]
* [G5.2, G5.2a, G5.2b, UL7.0] on line#3 of file [tests/testthat/test-cpr_make_pal.R]
* [G5.2, G5.2a, G5.2b, UL7.0] on line#3 of file [tests/testthat/test-cpr_rand_comm.R]
* [G5.2, G5.2a, G5.2b, UL7.0, G5.0, G2.11, G2.16, UL1.4] on line#56 of file [tests/testthat/test-cpr_rand_test.R]
* [UL1.2] on line#406 of file [tests/testthat/test-cpr_rand_test.R]
* [UL7.5, UL7.5a] on line#429 of file [tests/testthat/test-cpr_rand_test.R]
* [UL7.5, UL7.5a] on line#478 of file [tests/testthat/test-cpr_rand_test.R]
* [G5.4, G5.4b, G5.5] on line#537 of file [tests/testthat/test-cpr_rand_test.R]
* [UL1.3, UL7.3] on line#579 of file [tests/testthat/test-cpr_rand_test.R]
* [G1.1] on line#32 of file [./README.Rmd]
── @srrstatsNA standards (52 / 231):
* [G1.5, G1.6, G2.4, G2.4b, G2.4c, G2.4d, G2.4e, G2.5, G2.9, G2.10, G2.12, G2.14b, G2.14c, G3.1, G3.1a, G4.0, G5.4c, G5.6, G5.6a, G5.6b, G5.8, G5.8a, G5.8b, G5.8c, G5.8d, G5.9, G5.9a, G5.9b, G5.10, G5.11, G5.11a, G5.12, UL1.3a, UL1.4a, UL1.4b, UL2.1, UL2.2, UL2.3, UL3.0, UL3.1, UL3.2, UL3.3, UL4.0, UL4.1, UL4.2, UL4.3, UL4.4, UL6.0, UL6.1, UL6.2, UL7.2, UL7.4] on line#175 of file [R/srr-stats-standards.R]srrパッケージでスタンダードを
管理する
スタンダードがまだ実行されていなかったら、TODOとして報告される
## ──────────────────── rOpenSci Statistical Software Standards ───────────────────
##
##
##
## ── @srrstats standards (8 / 12):
##
## * [G1.1, G1.2, G1.3, G2.0, G2.1] in function 'test_fn()' on line#11 of file [R/test.R]
## * [RE2.2] on line#2 of file [tests/testthat/test-a.R]
## * [G2.3] in function 'test()' on line#6 of file [src/cpptest.cpp]
## * [G1.4] on line#17 of file [./README.Rmd]
##
##
##
## ── @srrstatsNA standards (1 / 12):
##
## * [RE3.3] on line#5 of file [R/srr-stats-standards.R]
##
##
##
## ── @srrstatsTODO standards (3 / 12):
##
## * [RE4.4] on line#14 of file [R/srr-stats-standards.R]
## * [RE1.1] on line#11 of file [R/test.R]
## * [G1.5] on line#17 of file [./README.Rmd]バッジ
 Bronze for software which is sufficiently or minimally compliant with standards to pass review.
Bronze for software which is sufficiently or minimally compliant with standards to pass review. Silver for software for which complies with more than a minimal set of applicable standards, and which extends beyond bronze in least one notable way.
Silver for software for which complies with more than a minimal set of applicable standards, and which extends beyond bronze in least one notable way. Gold for software which complies with all standards which reviewers have deemed potentially applicable.
Gold for software which complies with all standards which reviewers have deemed potentially applicable.
最後の一押し
ガイドブックやスタンダードを使いながらパッケージを書くだけでもかなり上達する
rOpenSciの強みは何よりも、コミュニティー
- 非常にRに詳しいメンバーがたくさんいて、すぐに(slackで)質問に答えてくれる
みんな様も是非試してみて下さい!