Content from はじめに
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- なぜ再現性にこだわる必要がありますか?
-
targetsは再現性の達成にどう役立ちますか?
Objectives
- 科学にとってなぜ再現性が重要なのかを説明しましょう。
- 再現性を高める
targetsの特徴を説明しましょう。
再現性とは?
再現性とは、他の人(未来の自分を含む)があなたの分析を再現できる能力のことです。
私たちは、科学的な分析結果が再現できる場合にのみ、その結果を信頼することができます。
しかし、再現性は二項対立的な概念(再現可能か再現不可能か)ではなく、再現性が低いものから高いものまでの尺度がある。
targetsは、あなたの分析をより再現性の高いものにするために大いに役立ちます。
再現性をさらに高めるために、Docker、conda、renvのようなツールを使ってコンピューティング環境を整えることもできますが、このワークショップではそれらをカバーする時間がありません。
targetsとは?
targetsはウィル・ランドーによって開発・管理されているRプログラミング言語用のワークフロー管理パッケージです。
targetsの主な特徴は以下の通りです:
- ワークフローの自動化
- ワークフロー・ステップのキャッシュ
- ワークフロー・ステップの一括作成
- ワークフローの段階での並列化
これにより、以下のことが可能になります:
- 別の作業をしてから元のプロジェクトに戻る際、混乱したり、何をしていたか思い出そうとしたりすることなく、すぐに中断したところから再開できます。
- ワークフローを変更し、変更の影響を受ける部分のみを再実行できます。
- 個々の機能を変更することなく、ワークフローを大幅に拡張できます。
… もちろん、これらはあなたの分析を他の人が再現するのにも役立ちます。
誰が targets を使うべきか?
targetsは決して唯一のワークフロー管理ソフトではありません。
似たようなツールは数多くあり、それぞれ機能や使用例が異なります。
例えば、 snakemakeはpython用の人気のあるワークフローツールで、makeはbashスクリプトを自動化するためのツールです。
targetsはR専用に設計されているので、Rを主に使う場合、あるいは使う予定がある場合は、targetsを使うのが最も理にかなっています。
他のツールでコーディングすることが多いのであれば、別の方法を検討したほうがいいかもしれません。
このワークショップのゴールは、Rで再現可能なデータ解析行うために**targetsの使用方法**を学ぶことです。
詳細情報
targetsは洗練されたパッケージであり、このワークショップではカバーしきれないほど学ぶべきことがたくさんあります。
targetsの旅を続けるためにお勧めのリソースをいくつか紹介します:
-
targetsの作者であるウィル・ランドーによるtargetsRパッケージ・ユーザーマニュアルは、targetsに真剣に興味を持つ人の必読書であす。 -
targetsの掲示板は、質問したり助けを求めたりするのに最適な場所です。 しかし、質問をする前に、必ず助けを求めることに関するポリシーを読みましょう。 -
targetsパッケージのウェブページには、すべてのtargetsの関数の説明が載っています。 -
tarchetypesパッケージのウェブページには、すべてのtarchetypesの関数の説明が載っています。tarchetypesはtargetsと一緒に使うことがほとんどなので、両方参照するのがおすすめです。 -
Reproducible
computation at scale in R with
targetsは、Kerasで顧客離れを分析するウィル・ランドーによるチュートリアルです。 -
targetsのREADMEに記載されている録画とプロジェクトの例。
サンプルデータセットについて
このワークショップでは、南極大陸のパーマー群島の島々で観察されたアデリー、ヒゲペンギン、ジェンツーペンギンの成鳥の採食行動に関する測定データセットの例を分析します。
データは palmerpenguins Rパッケージから入手できます。
?palmerpenguinsを実行すれば、データに関する詳細な情報を得ることができます。

分析の目的は、線形モデルを用いて嘴の長さと深さの関係を明らかにすることです。
このレッスンを通して徐々に分析を積み上げていきますが、最終版は https://github.com/joelnitta/penguins-targetsで見ることができます。
Content from 初めてのtargetsによるワークフロー
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- 解析を整理するためのベストプラクティスは?
-
_targets.Rファイルは何に使うのか? -
_targets.Rファイルの内容は? - ワークフローはどのように実行出来るのか?
Objectives
- RStudio でプロジェクトを作成する
-
_targets.Rファイルの目的を説明する - 基本的な
_targets.Rファイルを書く -
_targets.Rファイルを使用してワークフローを実行する
プロジェクトの作成
プロジェクトについて
targets
は、分析を整理するために「プロジェクト」という概念を採用しています。あるプロジェクトに必要なファイルはすべて、プロジェクトフォルダという1つのフォルダに収められます。
プロジェクトフォルダには、データ、コード、結果用のフォルダなど、整理のためのサブフォルダも追加可能です。
プロジェクトを使用することによって、別のプロジェクト(解析)から戻って来た際、迷わずに中断したところから再開することができます。 何かをやり始めたら完成するまで他のことに一切手を出さないなら、このような問題は起こりませんが、そのようなことはほとんどありません。 何か別の作業をしてから元のプロジェクトに戻ったとき、自分が何をしていたかを思い出すのは難しいですね(これは「コンテクスト・スイッチング」と呼ばれる現象です)。 標準化されたプロジェクト整理システムを使うことによって、混乱や時間のロスを減らすことができます。 つまり、再現性を高めているのです!
このワークショップではRStudioを使用します。RStudioは「プロジェクト」の概念と相性が良いからです。
RStudio でプロジェクトを作成する
RStudioを使って新しいプロジェクトを開始しましょう。
“File” をクリックし、“New Project” を選択します。
これにより、新規プロジェクトウィザードが開き、プロジェクトの設定に役立つメニューが表示されます。
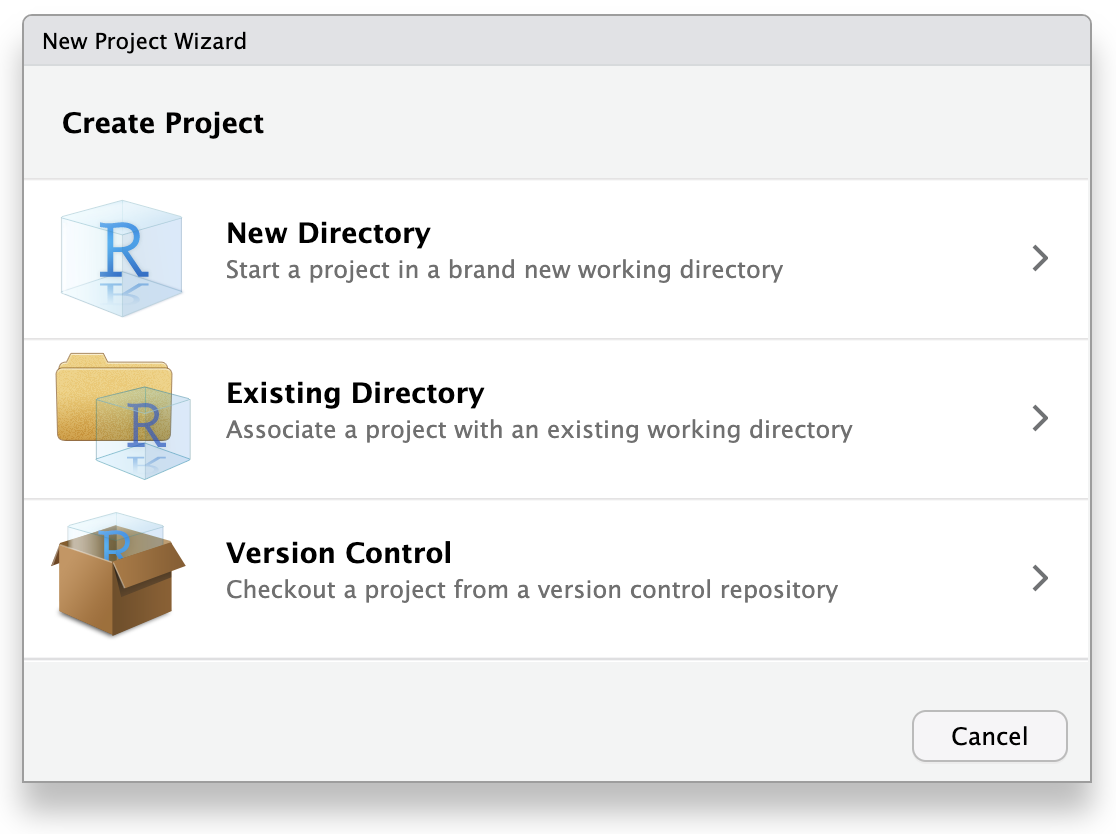
ウィザードでは、ゼロから新しいプロジェクトを作るので、最初のオプション「新規ディレクトリ」をクリックします。 Click “New Project” in the next menu. In “Directory name”, enter a name that helps you remember the purpose of the project, such as “targets-demo” (follow best practices for naming files and folders). Under “Create project as a subdirectory of…”, click the “Browse” button to select a directory to put the project. We recommend putting it on your Desktop so you can easily find it.
You can leave “Create a git repository” and “Use renv with this project” unchecked, but these are both excellent tools to improve reproducibility, and you should consider learning them and using them in the future, if you don’t already. They can be enabled at any later time, so you don’t need to worry about trying to use them immediately.
Once you work through these steps, your RStudio session should look like this:
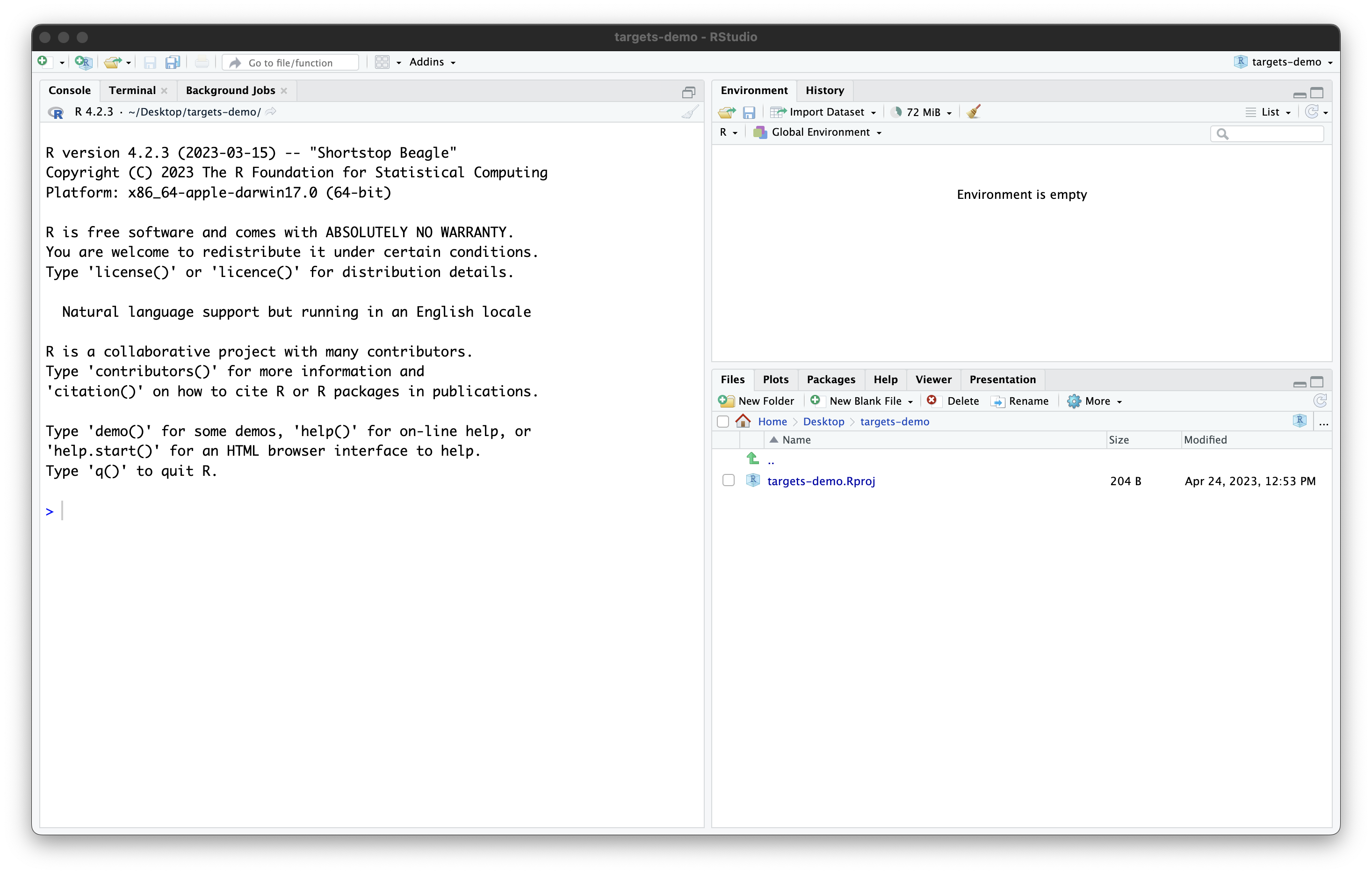
Our project now contains a single file, created by RStudio:
targets-demo.Rproj. You should not edit this file by hand.
Its purpose is to tell RStudio that this is a project folder and to
store some RStudio settings (if you use version-control software, it is
OK to commit this file). Also, you can open the project by double
clicking on the .Rproj file in your file explorer (try it
by quitting RStudio then navigating in your file browser to your
Desktop, opening the “targets-demo” folder, and double clicking
targets-demo.Rproj).
OK, now that our project is set up, we are ready to start using
targets!
Create a _targets.R file
Every targets project must include a special file,
called _targets.R in the main project folder (the “project
root”). The _targets.R file includes the specification of
the workflow: directions for R to run your analysis, kind of like a
recipe. By using the _targets.R file, you won’t have to
remember to run specific scripts in a certain order. Instead, R will do
it for you (more reproducibility points)!
Anatomy of a _targets.R file
We will now start to write a _targets.R file.
Fortunately, targets comes with a function to help us do
this.
In the R console, first load the targets package with
library(targets), then run the command
tar_script().
R
library(targets)
tar_script()
Nothing will happen in the console, but in the file viewer, you
should see a new file, _targets.R appear. Open it using the
File menu or by clicking on it.
We can see this default _targets.R file includes three
main parts:
- Loading packages with
library() - Defining a custom function with
function() - Defining a list with
list().
The last part, the list, is the most important part of the
_targets.R file. It defines the steps in the workflow. The
_targets.R file must always end with this list.
Furthermore, each item in the list is a call of the
tar_target() function. The first argument of
tar_target() is name of the target to build, and the second
argument is the command used to build it. Note that the name of the
target is unquoted, that is, it is written without any
surrounding quotation marks.
Set up _targets.R file to run example analysis
Background: non-targets version
We will use this template to start building our analysis of bill
shape in penguins. First though, to get familiar with the functions and
packages we’ll use, let’s run the code like you would in a “normal” R
script without using targets.
Recall that we are using the palmerpenguins R package to
obtain the data. This package actually includes two variations of the
dataset: one is an external CSV file with the raw data, and another is
the cleaned data loaded into R. In real life you are probably have
externally stored raw data, so let’s use the raw penguin
data as the starting point for our analysis too.
The path_to_file() function in
palmerpenguins provides the path to the raw data CSV file
(it is inside the palmerpenguins R package source code that
you downloaded to your computer when you installed the package).
R
library(palmerpenguins)
# Get path to CSV file
penguins_csv_file <- path_to_file("penguins_raw.csv")
penguins_csv_file
OUTPUT
[1] "/home/runner/.local/share/renv/cache/v5/R-4.3/x86_64-pc-linux-gnu/palmerpenguins/0.1.1/6c6861efbc13c1d543749e9c7be4a592/palmerpenguins/extdata/penguins_raw.csv"We will use the tidyverse set of packages for loading
and manipulating the data. We don’t have time to cover all the details
about using tidyverse now, but if you want to learn more
about it, please see the “Manipulating,
analyzing and exporting data with tidyverse” lesson.
Let’s load the data with read_csv().
R
library(tidyverse)
# Read CSV file into R
penguins_data_raw <- read_csv(penguins_csv_file)
penguins_data_raw
OUTPUT
Rows: 344 Columns: 17
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (9): studyName, Species, Region, Island, Stage, Individual ID, Clutch C...
dbl (7): Sample Number, Culmen Length (mm), Culmen Depth (mm), Flipper Leng...
date (1): Date Egg
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.OUTPUT
# A tibble: 344 × 17
studyName `Sample Number` Species Region Island Stage `Individual ID`
<chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 PAL0708 1 Adelie Penguin… Anvers Torge… Adul… N1A1
2 PAL0708 2 Adelie Penguin… Anvers Torge… Adul… N1A2
3 PAL0708 3 Adelie Penguin… Anvers Torge… Adul… N2A1
4 PAL0708 4 Adelie Penguin… Anvers Torge… Adul… N2A2
5 PAL0708 5 Adelie Penguin… Anvers Torge… Adul… N3A1
6 PAL0708 6 Adelie Penguin… Anvers Torge… Adul… N3A2
7 PAL0708 7 Adelie Penguin… Anvers Torge… Adul… N4A1
8 PAL0708 8 Adelie Penguin… Anvers Torge… Adul… N4A2
9 PAL0708 9 Adelie Penguin… Anvers Torge… Adul… N5A1
10 PAL0708 10 Adelie Penguin… Anvers Torge… Adul… N5A2
# ℹ 334 more rows
# ℹ 10 more variables: `Clutch Completion` <chr>, `Date Egg` <date>,
# `Culmen Length (mm)` <dbl>, `Culmen Depth (mm)` <dbl>,
# `Flipper Length (mm)` <dbl>, `Body Mass (g)` <dbl>, Sex <chr>,
# `Delta 15 N (o/oo)` <dbl>, `Delta 13 C (o/oo)` <dbl>, Comments <chr>We see the raw data has some awkward column names with spaces (these are hard to type out and can easily lead to mistakes in the code), and far more columns than we need. For the purposes of this analysis, we only need species name, bill length, and bill depth. In the raw data, the rather technical term “culmen” is used to refer to the bill.

Let’s clean up the data to make it easier to use for downstream analyses. We will also remove any rows with missing data, because this could cause errors for some functions later.
R
# Clean up raw data
penguins_data <- penguins_data_raw |>
# Rename columns for easier typing and
# subset to only the columns needed for analysis
select(
species = Species,
bill_length_mm = `Culmen Length (mm)`,
bill_depth_mm = `Culmen Depth (mm)`
) |>
# Delete rows with missing data
remove_missing(na.rm = TRUE)
penguins_data
OUTPUT
# A tibble: 342 × 3
species bill_length_mm bill_depth_mm
<chr> <dbl> <dbl>
1 Adelie Penguin (Pygoscelis adeliae) 39.1 18.7
2 Adelie Penguin (Pygoscelis adeliae) 39.5 17.4
3 Adelie Penguin (Pygoscelis adeliae) 40.3 18
4 Adelie Penguin (Pygoscelis adeliae) 36.7 19.3
5 Adelie Penguin (Pygoscelis adeliae) 39.3 20.6
6 Adelie Penguin (Pygoscelis adeliae) 38.9 17.8
7 Adelie Penguin (Pygoscelis adeliae) 39.2 19.6
8 Adelie Penguin (Pygoscelis adeliae) 34.1 18.1
9 Adelie Penguin (Pygoscelis adeliae) 42 20.2
10 Adelie Penguin (Pygoscelis adeliae) 37.8 17.1
# ℹ 332 more rowsThat’s better!
targets version
What does this look like using targets?
The biggest difference is that we need to put each step of the workflow into the list at the end.
We also define a custom function for the data cleaning step. That is because the list of targets at the end should look like a high-level summary of your analysis. You want to avoid lengthy chunks of code when defining the targets; instead, put that code in the custom functions. The other steps (setting the file path and loading the data) are each just one function call so there’s not much point in putting those into their own custom functions.
Finally, each step in the workflow is defined with the
tar_target() function.
R
library(targets)
library(tidyverse)
library(palmerpenguins)
clean_penguin_data <- function(penguins_data_raw) {
penguins_data_raw |>
select(
species = Species,
bill_length_mm = `Culmen Length (mm)`,
bill_depth_mm = `Culmen Depth (mm)`
) |>
remove_missing(na.rm = TRUE)
}
list(
tar_target(penguins_csv_file, path_to_file("penguins_raw.csv")),
tar_target(penguins_data_raw, read_csv(
penguins_csv_file, show_col_types = FALSE)),
tar_target(penguins_data, clean_penguin_data(penguins_data_raw))
)
I have set show_col_types = FALSE in
read_csv() because we know from the earlier code that the
column types were set correctly by default (character for species and
numeric for bill length and depth), so we don’t need to see the warning
it would otherwise issue.
Run the workflow
Now that we have a workflow, we can run it with the
tar_make() function. Try running it, and you should see
something like this:
R
tar_make()
OUTPUT
• start target penguins_csv_file
• built target penguins_csv_file [0.002 seconds]
• start target penguins_data_raw
• built target penguins_data_raw [0.095 seconds]
• start target penguins_data
• built target penguins_data [0.013 seconds]
• end pipeline [0.213 seconds]Congratulations, you’ve run your first workflow with
targets!
Key Points
- Projects help keep our analyses organized so we can easily re-run them later
- Use the RStudio Project Wizard to create projects
- The
_targets.Rfile is a special file that must be included in alltargetsprojects, and defines the worklow - Use
tar_script()to create a default_targets.Rfile - Use
tar_make()to run the workflow
Content from Loading Workflow Objects
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- ワークフローはどこで行われるのか?
- ワークフローによって構築されたオブジェクトを確認するには?
Objectives
- Explain where
targetsruns the workflow and why - Be able to load objects built by the workflow into your R session
ワークフローはどこで行われるのか?
So we just finished running our first workflow. Now you probably want
to look at its output. But, if we just call the name of the object (for
example, penguins_data), we get an error.
R
penguins_data
ERROR
Error in eval(expr, envir, enclos): object 'penguins_data' not foundWhere are the results of our workflow?
We don’t see the workflow results because targets
runs the workflow in a separate R session that we can’t
interact with. This is for reproducibility—the objects built by the
workflow should only depend on the code in your project, not any
commands you may have interactively given to R.
Fortunately, targets has two functions that can be used
to load objects built by the workflow into our current session,
tar_load() and tar_read(). Let’s see how these
work.
tar_load()
tar_load() loads an object built by the workflow into
the current session. Its first argument is the name of the object you
want to load. Let’s use this to load penguins_data and get
an overview of the data with summary().
R
tar_load(penguins_data)
summary(penguins_data)
OUTPUT
species bill_length_mm bill_depth_mm
Length:342 Min. :32.10 Min. :13.10
Class :character 1st Qu.:39.23 1st Qu.:15.60
Mode :character Median :44.45 Median :17.30
Mean :43.92 Mean :17.15
3rd Qu.:48.50 3rd Qu.:18.70
Max. :59.60 Max. :21.50 tar_load()は、その副作用、つまり、目的のオブジェクトを現在のRセッションにロードするために使用されることに注意してください。
It doesn’t actually return a value.
tar_read()
tar_read() is similar to tar_load() in that
it is used to retrieve objects built by the workflow, but unlike
tar_load(), it returns them directly as output.
Let’s try it with penguins_csv_file.
R
tar_read(penguins_csv_file)
OUTPUT
[1] "/home/runner/.local/share/renv/cache/v5/R-4.3/x86_64-pc-linux-gnu/palmerpenguins/0.1.1/6c6861efbc13c1d543749e9c7be4a592/palmerpenguins/extdata/penguins_raw.csv"We immediately see the contents of penguins_csv_file.
But it has not been loaded into the environment. If you try to run
penguins_csv_file now, you will get an error:
R
penguins_csv_file
ERROR
Error in eval(expr, envir, enclos): object 'penguins_csv_file' not foundWhen to use which function
tar_load() tends to be more useful when you want to load
objects and do things with them. tar_read() is more useful
when you just want to immediately inspect an object.
The targets cache
If you close your R session, then re-start it and use
tar_load() or tar_read(), you will notice that
it can still load the workflow objects. In other words, the workflow
output is saved across R sessions. How is this
possible?
You may have noticed a new folder has appeared in your project,
called _targets. This is the targets
cache. It contains all of the workflow output; that is how we
can load the targets built by the workflow even after quitting then
restarting R.
You should not edit the contents of the cache by hand (with one exception). Doing so would make your analysis non-reproducible.
The one exception to this rule is a special subfolder called
_targets/user. This folder does not exist by default. You
can create it if you want, and put whatever you want inside.
Generally, _targets/user is a good place to store files
that are not code, like data and output.
Note that if you don’t have anything in _targets/user
that you need to keep around, it is possible to “reset” your workflow by
simply deleting the entire _targets folder. Of course, this
means you will need to run everything over again, so don’t do this
lightly!
Content from The Workflow Lifecycle
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- What happens if we re-run a workflow?
- How does
targetsknow what steps to re-run? - How can we inspect the state of the workflow?
Objectives
- Explain how
targetshelps increase efficiency - Be able to inspect a workflow to see what parts are outdated
Re-running the workflow
One of the features of targets is that it maximizes
efficiency by only running the parts of the workflow that need to be
run.
This is easiest to understand by trying it yourself. Let’s try running the workflow again:
R
tar_make()
OUTPUT
✔ skip target penguins_csv_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
✔ skip pipeline [0.078 seconds]Remember how the first time we ran the pipeline, targets
printed out a list of each target as it was being built?
This time, it tells us it is skipping those targets; they have already been built, so there’s no need to run that code again.
Remember, the fastest code is the code you don’t have to run!
Re-running the workflow after modification
What happens when we change one part of the workflow then run it again?
Say that we decide the species names should be shorter. Right now they include the common name and the scientific name, but we really only need the first part of the common name to distinguish them.
Edit _targets.R so that the
clean_penguin_data() function looks like this:
R
clean_penguin_data <- function(penguins_data_raw) {
penguins_data_raw |>
select(
species = Species,
bill_length_mm = `Culmen Length (mm)`,
bill_depth_mm = `Culmen Depth (mm)`
) |>
remove_missing(na.rm = TRUE) |>
# Split "species" apart on spaces, and only keep the first word
separate(species, into = "species", extra = "drop")
}
Then run it again.
R
tar_make()
OUTPUT
✔ skip target penguins_csv_file
✔ skip target penguins_data_raw
• start target penguins_data
• built target penguins_data [0.025 seconds]
• end pipeline [0.12 seconds]What happened?
This time, it skipped penguins_csv_file and
penguins_data_raw and only ran
penguins_data.
Of course, since our example workflow is so short we don’t even notice the amount of time saved. But imagine using this in a series of computationally intensive analysis steps. The ability to automatically skip steps results in a massive increase in efficiency.
With tar_read(penguins_data) or by running
tar_load(penguins_data) followed by
penguins_data.
Under the hood
How does targets keep track of which targets are
up-to-date vs. outdated?
For each target in the workflow (items in the list at the end of the
_targets.R file) and any custom functions used in the
workflow, targets calculates a hash value,
or unique combination of letters and digits that represents an object in
the computer’s memory. You can think of the hash value (or “hash” for
short) as a unique fingerprint for a target or
function.
The first time your run tar_make(), targets
calculates the hashes for each target and function as it runs the code
and stores them in the targets cache (the _targets folder).
Then, for each subsequent call of tar_make(), it calculates
the hashes again and compares them to the stored values. It detects
which have changed, and this is how it knows which targets are out of
date.
This information is used in combination with the dependency relationships (in other words, how each target depends on the others) to re-run the workflow in the most efficient way possible: code is only run for targets that need to be re-built, and others are skipped.
Visualizing the workflow
Typically, you will be making edits to various places in your code, adding new targets, and running the workflow periodically. It is good to be able to visualize the state of the workflow.
This can be done with tar_visnetwork()
R
tar_visnetwork()
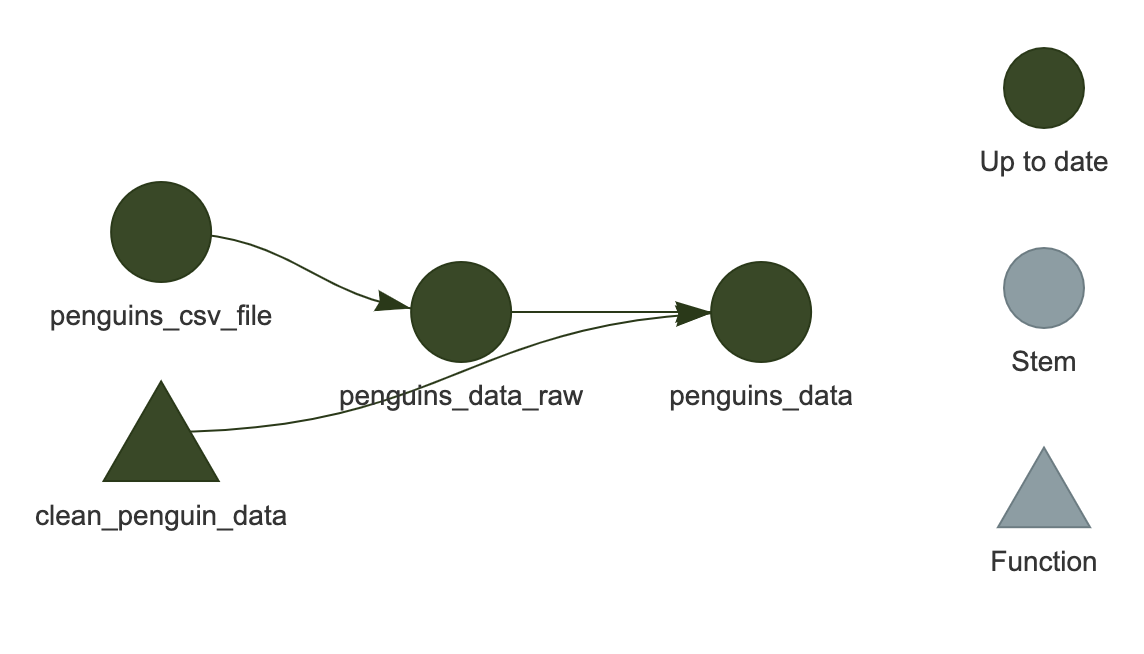
You should see the network show up in the plot area of RStudio.
It is an HTML widget, so you can zoom in and out (this isn’t important for the current example since it is so small, but is useful for larger, “real-life” workflows).
Here, we see that all of the targets are dark green, indicating that they are up-to-date and would be skipped if we were to run the workflow again.
Light blue indicates the target is out of date.
Depending on how you modified the code, any or all of the targets may now be light blue.
‘Outdated’ does not always mean ‘will be run’
Just because a target appears as light blue (is “outdated”) in the network visualization, this does not guarantee that it will be re-built during the next run. Rather, it means that at least of one the targets that it depends on has changed.
For example, if the workflow state looked like this:
A -> B* -> C -> D
where the * indicates that B has changed
compared to the last time the workflow was run, the network
visualization will show B, C, and
D all as light blue.
But if re-running the workflow results in the exact same value for
C as before, D will not be re-run (will be
“skipped”).
Most of the time, a single change will cascade to the rest of the
downstream targets and cause them to be re-built, but this is not always
the case. targets has no way of knowing ahead of time what
the actual output will be, so it cannot provide a network visualization
that completely predicts the future!
Other ways to check workflow status
The visualization is very useful, but sometimes you may be working on a server that doesn’t provide graphical output, or you just want a quick textual summary of the workflow. There are some other useful functions that can do that.
tar_outdated() lists only the outdated targets; that is,
targets that will be built during the next run, or depend on such a
target. If everything is up to date, it will return a zero-length
character vector (character(0)).
R
tar_outdated()
OUTPUT
character(0)tar_progress() shows the current status of the workflow
as a dataframe. You may find it helpful to further manipulate the
dataframe to obtain useful summaries of the workflow, for example using
dplyr (such data manipulation is beyond the scope of this
lesson but the instructor may demonstrate its use).
R
tar_progress()
OUTPUT
# A tibble: 3 × 2
name progress
<chr> <chr>
1 penguins_csv_file skipped
2 penguins_data_raw skipped
3 penguins_data built Granular control of targets
It is possible to only make a particular target instead of running the entire workflow.
To do this, type the name of the target you wish to build after
tar_make() (note that any targets required by the one you
specify will also be built). For example,
tar_make(penguins_data_raw) would only
build penguins_data_raw, not
penguins_data.
Furthermore, if you want to manually “reset” a target and make it
appear out-of-date, you can do so with tar_invalidate().
This means that target (and any that depend on it) will be re-run next
time.
Let’s give this a try. Remember that our pipeline is currently up to
date, so tar_make() will skip everything:
R
tar_make()
OUTPUT
✔ skip target penguins_csv_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
✔ skip pipeline [0.073 seconds]Let’s invalidate penguins_data and run it again:
R
tar_invalidate(penguins_data)
tar_make()
OUTPUT
✔ skip target penguins_csv_file
✔ skip target penguins_data_raw
• start target penguins_data
• built target penguins_data [0.036 seconds]
• end pipeline [0.122 seconds]If you want to reset everything and start fresh, you
can use tar_invalidate(everything())
(tar_invalidate() accepts
tidyselect expressions to specify target names).
Caution should be exercised when using granular
methods like this, though, since you may end up with your workflow in an
unexpected state. The surest way to maintain an up-to-date workflow is
to run tar_make() frequently.
How this all works in practice
In practice, you will likely be switching between running the
workflow with tar_make(), loading the targets you built
with tar_load(), and editing your custom functions by
running code in an interactive R session. It takes some time to get used
to it, but soon you will feel that your code isn’t “real” until it is
embedded in a targets workflow.
Key Points
-
targetsonly runs the steps that have been affected by a change to the code -
tar_visnetwork()shows the current state of the workflow as a network -
tar_progress()shows the current state of the workflow as a data frame -
tar_outdated()lists outdated targets -
tar_invalidate()can be used to invalidate (re-run) specific targets
Content from Best Practices for targets Project Organization
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- What are best practices for organizing
targetsprojects? - How does the organization of a
targetsworkflow differ from a script-based analysis?
Objectives
- Explain how to organize
targetsprojects for maximal reproducibility - Understand how to use functions in the context of
targets
A simpler way to write workflow plans
The default way to specify targets in the plan is with the
tar_target() function. But this way of writing plans can be
a bit verbose.
There is an alternative provided by the tarchetypes
package, also written by the creator of targets, Will
Landau.
The purpose of the tarchetypes is to provide various
shortcuts that make writing targets pipelines easier. We
will introduce just one for now, tar_plan(). This is used
in place of list() at the end of the
_targets.R script. By using tar_plan(),
instead of specifying targets with tar_target(), we can use
a syntax like this: target_name = target_command.
Let’s edit the penguins workflow to use the tar_plan()
syntax:
R
library(targets)
library(tarchetypes)
library(palmerpenguins)
library(tidyverse)
clean_penguin_data <- function(penguins_data_raw) {
penguins_data_raw |>
select(
species = Species,
bill_length_mm = `Culmen Length (mm)`,
bill_depth_mm = `Culmen Depth (mm)`
) |>
remove_missing(na.rm = TRUE) |>
# Split "species" apart on spaces, and only keep the first word
separate(species, into = "species", extra = "drop")
}
tar_plan(
penguins_csv_file = path_to_file("penguins_raw.csv"),
penguins_data_raw = read_csv(penguins_csv_file, show_col_types = FALSE),
penguins_data = clean_penguin_data(penguins_data_raw)
)
I think it is easier to read, do you?
Notice that tar_plan() does not mean you have to write
all targets this way; you can still use the
tar_target() format within tar_plan(). That is
because =, while short and easy to read, does not provide
all of the customization that targets is capable of. This
doesn’t matter so much for now, but it will become important when you
start to create more advanced targets workflows.
Organizing files and folders
So far, we have been doing everything with a single
_targets.R file. This is OK for a small workflow, but does
not work very well when the workflow gets bigger. There are better ways
to organize your code.
First, let’s create a directory called R to store R code
other than _targets.R (remember,
_targets.R must be placed in the overall project directory,
not in a subdirectory). Create a new R file in R/ called
functions.R. This is where we will put our custom
functions. Let’s go ahead and put clean_penguin_data() in
there now and save it.
Similarly, let’s put the library() calls in their own
script in R/ called packages.R (this isn’t the
only way to do it though; see the “Managing
Packages” episode for alternative approaches).
We will also need to modify our _targets.R script to
call these scripts with source:
R
source("R/packages.R")
source("R/functions.R")
tar_plan(
penguins_csv_file = path_to_file("penguins_raw.csv"),
penguins_data_raw = read_csv(penguins_csv_file, show_col_types = FALSE),
penguins_data = clean_penguin_data(penguins_data_raw)
)
Now _targets.R is much more streamlined: it is focused
just on the workflow and immediately tells us what happens in each
step.
Finally, let’s make some directories for storing data and
output—files that are not code. Create a new directory inside the
targets cache called user: _targets/user.
Within user, create two more directories, data
and results. (If you use version control, you will probably
want to ignore the _targets directory).
A word about functions
We mentioned custom functions earlier in the lesson, but this is an
important topic that deserves further clarification. If you are used to
analyzing data in R with a series of scripts instead of a single
workflow like targets, you may not write many functions
(using the function() function).
This is a major difference from targets. It would be
quite difficult to write an efficient targets pipeline
without the use of custom functions, because each target you build has
to be the output of a single command.
We don’t have time in this curriculum to cover how to write functions in R, but the Software Carpentry lesson is recommended for reviewing this topic.
Another major difference is that each target must have a unique name. You may be used to writing code that looks like this:
R
# Store a person's height in cm, then convert to inches
height <- 160
height <- height / 2.54
You would get an error if you tried to run the equivalent targets pipeline:
R
tar_plan(
height = 160,
height = height / 2.54
)
ERROR
Error:
! Error running targets::tar_make()
Error messages: targets::tar_meta(fields = error, complete_only = TRUE)
Debugging guide: https://books.ropensci.org/targets/debugging.html
How to ask for help: https://books.ropensci.org/targets/help.html
Last error: duplicated target names: heightA major part of working with targets pipelines
is writing custom functions that are the right size. They
should not be so small that each is just a single line of code; this
would make your pipeline difficult to understand and be too difficult to
maintain. On the other hand, they should not be so big that each has
large numbers of inputs and is thus overly sensitive to changes.
Striking this balance is more of art than science, and only comes with practice. I find a good rule of thumb is no more than three inputs per target.
Content from Managing Packages
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How should I manage packages for my
targetsproject?
Objectives
- Demonstrate best practices for managing packages
Loading packages
Almost every R analysis relies on packages for functions beyond those available in base R.
There are three main ways to load packages in targets
workflows.
Method 1: library()
This is the method you are almost certainly more familiar with, and is the method we have been using by default so far.
Like any other R script, include library() calls near
the top of the _targets.R script. Alternatively (and as the
recommended best practice for project
organization), you can put all of the library() calls in a
separate script—this is typically called packages.R and
stored in the R/ directory of your project.
The potential downside to this approach is that if you have a long
list of packages to load, certain functions like
tar_visnetwork(), tar_outdated(), etc., may
take an unnecessarily long time to run because they have to load all the
packages, even though they don’t necessarily use them.
Method 2: tar_option_set()
In this method, use the tar_option_set() function in
_targets.R to specify the packages to load when running the
workflow.
This will be demonstrated using the pre-cleaned dataset from the
palmerpenguins package. Let’s say we want to filter it down
to just data for the Adelie penguin.
Save your progress
You can only have one active _targets.R file at a time
in a given project.
We are about to create a new _targets.R file, but you
probably don’t want to lose your progress in the one we have been
working on so far (the penguins bill analysis). You can temporarily
rename that one to something like _targets_old.R so that
you don’t overwrite it with the new example _targets.R file
below. Then, rename them when you are ready to work on it again.
This is what using the tar_option_set() method looks
like:
R
library(targets)
library(tarchetypes)
tar_option_set(packages = c("dplyr", "palmerpenguins"))
tar_plan(
adelie_data = filter(penguins, species == "Adelie")
)
OUTPUT
• start target adelie_data
• built target adelie_data [0.029 seconds]
• end pipeline [0.104 seconds]This method gets around the slow-downs that may sometimes be experienced with Method 1.
Method 3: packages argument of
tar_target()
The main function for defining targets, tar_target()
includes a packages argument that will load the specified
packages only for that target.
Here is how we could use this method, modified from the same example as above.
R
library(targets)
library(tarchetypes)
tar_plan(
tar_target(
adelie_data,
filter(penguins, species == "Adelie"),
packages = c("dplyr", "palmerpenguins")
)
)
OUTPUT
• start target adelie_data
• built target adelie_data [0.03 seconds]
• end pipeline [0.103 seconds]This can be more memory efficient in some cases than loading all packages, since not every target is always made during a typical run of the workflow. But, it can be tedious to remember and specify packages needed on a per-target basis.
One more option
Another alternative that does not actually involve loading packages
is to specify the package associated with each function by using the
:: notation, for example, dplyr::mutate().
This means you can avoid loading packages
altogether.
Here is how to write the plan using this method:
R
library(targets)
library(tarchetypes)
tar_plan(
adelie_data = dplyr::filter(palmerpenguins::penguins, species == "Adelie")
)
OUTPUT
• start target adelie_data
• built target adelie_data [0.021 seconds]
• end pipeline [0.095 seconds]The benefits of this approach are that the origins of all functions is explicit, so you could browse your code (for example, by looking at its source in GitHub), and immediately know where all the functions come from. The downside is that it is rather verbose because you need to type the package name every time you use one of its functions.
Which is the right way?
There is no “right” answer about how to load packages—it is a matter of what works best for your particular situation.
Often a reasonable approach is to load your most commonly used
packages with library() (such as tidyverse) in
packages.R, then use :: notation for less
frequently used functions whose origins you may otherwise forget.
Maintaining package versions
Tracking of custom functions vs. functions from packages
A critical thing to understand about targets is that
it only tracks custom functions and targets, not
functions provided by packages.
However, the content of packages can change, and packages typically get updated on a regular basis. The output of your workflow may depend not only on the packages you use, but their versions.
Therefore, it is a good idea to track package versions.
About renv
Fortunately, you don’t have to do this by hand: there are R packages
available that can help automate this process. We recommend renv, but there are
others available as well (e.g., groundhog). We don’t have the time to
cover detailed usage of renv in this lesson. To get started
with renv, see the “Introduction
to renv” vignette.
You can generally use renv the same way you would for a
targets project as any other R project. However, there is
one exception: if you load packages using tar_option_set()
or the packages argument of tar_target() (Method 2 or Method 3,
respectively), renv will not detect them (because it
expects packages to be loaded with library(),
require(), etc.).
The solution in this case is to use the tar_renv()
function. This will write a separate file with
library() calls for each package used in the workflow so
that renv will properly detect them.
Selective tracking of functions from packages
Because targets doesn’t track functions from packages,
if you update a package and the contents of one of its functions
changes, targets will not re-build the target that
was generated by that function.
However, it is possible to change this behavior on a per-package
basis. This is best done only for a small number of packages, since
adding too many would add too much computational overhead to
targets when it has to calculate dependencies. For example,
you may want to do this if you are using your own custom package that
you update frequently.
The way to do so is by using tar_option_set(),
specifying the same package name in both
packages and imports. Here is a modified
version of the earlier code that demonstrates this for
dplyr and palmerpenguins.
R
library(targets)
library(tarchetypes)
tar_option_set(
packages = c("dplyr", "palmerpenguins"),
imports = c("dplyr", "palmerpenguins")
)
tar_plan(
adelie_data = filter(penguins, species == "Adelie")
)
If we were to re-install either dplyr or
palmerpenguins and one of the functions used from those in
the pipeline changes (for example, filter()), any target
depending on that function will be rebuilt.
Resolving namespace conflicts
There is one final best-practice to mention related to packages: resolving namespace conflicts.
“Namespace” refers to the idea that a certain set of unique names are only unique within a particular context. For example, all the function names of a package have to be unique, but only within that package. Function names could be duplicated across packages.
As you may imagine, this can cause confusion. For example, the
filter() function appears in both the stats
package and the dplyr package, but does completely
different things in each. This is a namespace conflict:
how do we know which filter() we are talking about?
The conflicted package can help prevent such confusion
by stopping you if you try to use an ambiguous function, and help you be
explicit about which package to use. We don’t have time to cover the
details here, but you can read more about how to use
conflicted at its website.
When you use conflicted, you will typically run a series
of commands to explicitly resolve namespace conflicts, like
conflicts_prefer(dplyr::filter) (this would tell R that we
want to use filter from dplyr, not
stats).
To use this in a targets workflow, you should put all
calls to conflicts_prefer in a special file called
.Rprofile that is located in the main folder of your
project. This will ensure that the conflicts are always resolved for
each target.
The recommended way to edit your .Rprofile is to use
usethis::edit_r_profile("project"). This will open
.Rprofile in your editor, where you can edit it and save
it.
For example, your .Rprofile could include this:
R
library(conflicted)
conflicts_prefer(dplyr::filter)
Note that you don’t need to run source() to run the code
in .Rprofile. It will always get run at the start of each R
session automatically.
Content from Working with External Files
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How can we load external data?
Objectives
- Be able to load external data into a workflow
- Configure the workflow to rerun if the contents of the external data change
Treating external files as a dependency
Almost all workflows will start by importing data, which is typically stored as an external file.
As a simple example, let’s create an external data file in RStudio
with the “New File” menu option. Enter a single line of text, “Hello
World” and save it as “hello.txt” text file in
_targets/user/data/.
We will read in the contents of this file and store it as
some_data in the workflow by writing the following plan and
running tar_make():
Save your progress
You can only have one active _targets.R file at a time
in a given project.
We are about to create a new _targets.R file, but you
probably don’t want to lose your progress in the one we have been
working on so far (the penguins bill analysis). You can temporarily
rename that one to something like _targets_old.R so that
you don’t overwrite it with the new example _targets.R file
below. Then, rename them when you are ready to work on it again.
R
library(targets)
library(tarchetypes)
tar_plan(
some_data = readLines("_targets/user/data/hello.txt")
)
OUTPUT
• start target some_data
• built target some_data [0.002 seconds]
• end pipeline [0.083 seconds]If we inspect the contents of some_data with
tar_read(some_data), it will contain the string
"Hello World" as expected.
Now say we edit “hello.txt”, perhaps add some text: “Hello World. How are you?”. Edit this in the RStudio text editor and save it. Now run the pipeline again.
R
library(targets)
library(tarchetypes)
tar_plan(
some_data = readLines("_targets/user/data/hello.txt")
)
OUTPUT
✔ skip target some_data
✔ skip pipeline [0.065 seconds]The target some_data was skipped, even though the
contents of the file changed.
That is because right now, targets is only tracking the
name of the file, not its contents. We need to use a
special function for that, tar_file() from the
tarchetypes package. tar_file() will calculate
the “hash” of a file—a unique digital signature that is determined by
the file’s contents. If the contents change, the hash will change, and
this will be detected by targets.
R
library(targets)
library(tarchetypes)
tar_plan(
tar_file(data_file, "_targets/user/data/hello.txt"),
some_data = readLines(data_file)
)
OUTPUT
• start target data_file
• built target data_file [0.001 seconds]
• start target some_data
• built target some_data [0 seconds]
• end pipeline [0.092 seconds]This time we see that targets does successfully re-build
some_data as expected.
A shortcut (or, About target factories)
However, also notice that this means we need to write two targets
instead of one: one target to track the contents of the file
(data_file), and one target to store what we load from the
file (some_data).
It turns out that this is a common pattern in targets
workflows, so tarchetypes provides a shortcut to express
this more concisely, tar_file_read().
R
library(targets)
library(tarchetypes)
tar_plan(
tar_file_read(
hello,
"_targets/user/data/hello.txt",
readLines(!!.x)
)
)
Let’s inspect this pipeline with tar_manifest():
R
tar_manifest()
OUTPUT
# A tibble: 2 × 2
name command
<chr> <chr>
1 hello_file "\"_targets/user/data/hello.txt\""
2 hello "readLines(hello_file)" Notice that even though we only specified one target in the pipeline
(hello, with tar_file_read()), the pipeline
actually includes two targets, hello_file
and hello.
That is because tar_file_read() is a special function
called a target factory, so-called because it makes
multiple targets at once. One of the main purposes of
the tarchetypes package is to provide target factories to
make writing pipelines easier and less error-prone.
Non-standard evaluation
What is the deal with the !!.x? That may look unfamiliar
even if you are used to using R. It is known as “non-standard
evaluation,” and gets used in some special contexts. We don’t have time
to go into the details now, but just remember that you will need to use
this special notation with tar_file_read(). If you forget
how to write it (this happens frequently!) look at the examples in the
help file by running ?tar_file_read.
Other data loading functions
Although we used readLines() as an example here, you can
use the same pattern for other functions that load data from external
files, such as readr::read_csv(),
xlsx::read_excel(), and others (for example,
read_csv(!!.x), read_excel(!!.x), etc.).
This is generally recommended so that your pipeline stays up to date with your input data.
R
source("R/packages.R")
source("R/functions.R")
tar_plan(
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
penguins_data = clean_penguin_data(penguins_data_raw)
)
OUTPUT
• start target penguins_data_raw_file
• built target penguins_data_raw_file [0.002 seconds]
• start target penguins_data_raw
• built target penguins_data_raw [0.098 seconds]
• start target penguins_data
• built target penguins_data [0.023 seconds]
• end pipeline [0.217 seconds]Writing out data
Writing to files is similar to loading in files: we will use the
tar_file() function. There is one important caveat: in this
case, the second argument of tar_file() (the command to
build the target) must return the path to the file. Not
all functions that write files do this (some return nothing; these treat
the output file is a side-effect of running the function), so you may
need to define a custom function that writes out the file and then
returns its path.
Let’s do this for writeLines(), the R function that
writes character data to a file. Normally, its output would be
NULL (nothing), as we can see here:
R
x <- writeLines("some text", "test.txt")
x
OUTPUT
NULLHere is our modified function that writes character data to a file
and returns the name of the file (the ... means “pass the
rest of these arguments to writeLines()”):
R
write_lines_file <- function(text, file, ...) {
writeLines(text = text, con = file, ...)
file
}
Let’s try it out:
R
x <- write_lines_file("some text", "test.txt")
x
OUTPUT
[1] "test.txt"We can now use this in a pipeline. For example let’s change the text to upper case then write it out again:
R
library(targets)
library(tarchetypes)
source("R/functions.R")
tar_plan(
tar_file_read(
hello,
"_targets/user/data/hello.txt",
readLines(!!.x)
),
hello_caps = toupper(hello),
tar_file(
hello_caps_out,
write_lines_file(hello_caps, "_targets/user/results/hello_caps.txt")
)
)
OUTPUT
• start target hello_file
• built target hello_file [0.002 seconds]
• start target hello
• built target hello [0 seconds]
• start target hello_caps
• built target hello_caps [0 seconds]
• start target hello_caps_out
• built target hello_caps_out [0.001 seconds]
• end pipeline [0.098 seconds]Take a look at hello_caps.txt in the
results folder and verify it is as you expect.
targets detects that hello_caps_out has
changed (is “invalidated”), and re-runs the code to make it, thus
writing out hello_caps.txt to results
again.
So this way of writing out results makes your pipeline more robust:
we have a guarantee that the contents of the file in
results are generated solely by the code in your plan.
Key Points
-
tarchetypes::tar_file()tracks the contents of a file - Use
tarchetypes::tar_file_read()in combination with data loading functions likeread_csv()to keep the pipeline in sync with your input data - Use
tarchetypes::tar_file()in combination with a function that writes to a file and returns its path to write out data
Content from Branching
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How can we specify many targets without typing everything out?
Objectives
- Be able to specify targets using branching
Why branching?
One of the major strengths of targets is the ability to
define many targets from a single line of code (“branching”). This not
only saves you typing, it also reduces the risk of
errors since there is less chance of making a typo.
Types of branching
There are two types of branching, dynamic branching
and static branching. “Branching” refers to the idea
that you can provide a single specification for how to make targets (the
“pattern”), and targets generates multiple targets from it
(“branches”). “Dynamic” means that the branches that result from the
pattern do not have to be defined ahead of time—they are a dynamic
result of the code.
In this workshop, we will only cover dynamic branching since it is
generally easier to write (static branching requires use of meta-programming,
an advanced topic). For more information about each and when you might
want to use one or the other (or some combination of the two), see the
targets package manual.
Example without branching
To see how this works, let’s continue our analysis of the
palmerpenguins dataset.
Our hypothesis is that bill depth decreases with bill length. We will test this hypothesis with a linear model.
For example, this is a model of bill depth dependent on bill length:
R
lm(bill_depth_mm ~ bill_length_mm, data = penguins_data)
We can add this to our pipeline. We will call it the
combined_model because it combines all the species together
without distinction:
R
source("R/packages.R")
source("R/functions.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build model
combined_model = lm(
bill_depth_mm ~ bill_length_mm,
data = penguins_data
)
)
OUTPUT
✔ skip target penguins_data_raw_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
• start target combined_model
• built target combined_model [0.026 seconds]
• end pipeline [0.123 seconds]Let’s have a look at the model. We will use the glance()
function from the broom package. Unlike base R
summary(), this function returns output as a tibble (the
tidyverse equivalent of a dataframe), which as we will see later is
quite useful for downstream analyses.
R
library(broom)
tar_load(combined_model)
glance(combined_model)
OUTPUT
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 0.0552 0.0525 1.92 19.9 0.0000112 1 -708. 1422. 1433. 1256. 340 342Notice the small P-value. This seems to indicate that the model is highly significant.
But wait a moment… is this really an appropriate model? Recall that there are three species of penguins in the dataset. It is possible that the relationship between bill depth and length varies by species.
We should probably test some alternative models. These could include models that add a parameter for species, or add an interaction effect between species and bill length.
Now our workflow is getting more complicated. This is what a workflow
for such an analysis might look like without branching
(make sure to add library(broom) to
packages.R):
R
source("R/packages.R")
source("R/functions.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
combined_model = lm(
bill_depth_mm ~ bill_length_mm,
data = penguins_data
),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species,
data = penguins_data
),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species,
data = penguins_data
),
# Get model summaries
combined_summary = glance(combined_model),
species_summary = glance(species_model),
interaction_summary = glance(interaction_model)
)
OUTPUT
✔ skip target penguins_data_raw_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
✔ skip target combined_model
• start target interaction_model
• built target interaction_model [0.004 seconds]
• start target species_model
• built target species_model [0.01 seconds]
• start target combined_summary
• built target combined_summary [0.008 seconds]
• start target interaction_summary
• built target interaction_summary [0.002 seconds]
• start target species_summary
• built target species_summary [0.003 seconds]
• end pipeline [0.135 seconds]Let’s look at the summary of one of the models:
R
tar_read(species_summary)
OUTPUT
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 0.769 0.767 0.953 375. 3.65e-107 3 -467. 944. 963. 307. 338 342So this way of writing the pipeline works, but is repetitive: we have
to call glance() each time we want to obtain summary
statistics for each model. Furthermore, each summary target
(combined_summary, etc.) is explicitly named and typed out
manually. It would be fairly easy to make a typo and end up with the
wrong model being summarized.
Example with branching
First attempt
Let’s see how to write the same plan using dynamic branching:
R
source("R/packages.R")
source("R/functions.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance(models[[1]]),
pattern = map(models)
)
)
What is going on here?
First, let’s look at the messages provided by
tar_make().
OUTPUT
✔ skip target penguins_data_raw_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
• start target models
• built target models [0.006 seconds]
• start branch model_summaries_5ad4cec5
• built branch model_summaries_5ad4cec5 [0.008 seconds]
• start branch model_summaries_c73912d5
• built branch model_summaries_c73912d5 [0.002 seconds]
• start branch model_summaries_91696941
• built branch model_summaries_91696941 [0.003 seconds]
• built pattern model_summaries
• end pipeline [0.14 seconds]There is a series of smaller targets (branches) that are each named
like model_summaries_5ad4cec5, then one overall
model_summaries target. That is the result of specifying
targets using branching: each of the smaller targets are the “branches”
that comprise the overall target. Since targets has no way
of knowing ahead of time how many branches there will be or what they
represent, it names each one using this series of numbers and letters
(the “hash”). targets builds each branch one at a time,
then combines them into the overall target.
Next, let’s look in more detail about how the workflow is set up, starting with how we defined the models:
R
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),Unlike the non-branching version, we defined the models in a
list (instead of one target per model). This is because dynamic
branching is similar to the base::apply() or purrrr::map()
method of looping: it applies a function to each element of a list. So
we need to prepare the input for looping as a list.
Next, take a look at the command to build the target
model_summaries.
R
# Get model summaries
tar_target(
model_summaries,
glance(models[[1]]),
pattern = map(models)
)
As before, the first argument is the name of the target to build, and the second is the command to build it.
Here, we apply the glance() function to each element of
models (the [[1]] is necessary because when
the function gets applied, each element is actually a nested list, and
we need to remove one layer of nesting).
Finally, there is an argument we haven’t seen before,
pattern, which indicates that this target should be built
using dynamic branching. map means to apply the command to
each element of the input list (models) sequentially.
Now that we understand how the branching workflow is constructed, let’s inspect the output:
R
tar_read(model_summaries)
OUTPUT
# A tibble: 3 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 0.0552 0.0525 1.92 19.9 1.12e- 5 1 -708. 1422. 1433. 1256. 340 342
2 0.769 0.767 0.953 375. 3.65e-107 3 -467. 944. 963. 307. 338 342
3 0.770 0.766 0.955 225. 8.52e-105 5 -466. 947. 974. 306. 336 342The model summary statistics are all included in a single dataframe.
But there’s one problem: we can’t tell which row came from which model! It would be unwise to assume that they are in the same order as the list of models.
This is due to the way dynamic branching works: by default, there is no information about the provenance of each target preserved in the output.
How can we fix this?
Second attempt
The key to obtaining useful output from branching pipelines is to
include the necessary information in the output of each individual
branch. Here, we want to know the kind of model that corresponds to each
row of the model summaries. To do that, we need to write a
custom function. You will need to write custom
functions frequently when using targets, so it’s good to
get used to it!
Here is the function. Save this in R/functions.R:
R
glance_with_mod_name <- function(model_in_list) {
model_name <- names(model_in_list)
model <- model_in_list[[1]]
glance(model) |>
mutate(model_name = model_name)
}
Our new pipeline looks almost the same as before, but this time we
use the custom function instead of glance().
R
source("R/functions.R")
source("R/packages.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance_with_mod_name(models),
pattern = map(models)
)
)
OUTPUT
✔ skip target penguins_data_raw_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
✔ skip target models
• start branch model_summaries_5ad4cec5
• built branch model_summaries_5ad4cec5 [0.029 seconds]
• start branch model_summaries_c73912d5
• built branch model_summaries_c73912d5 [0.006 seconds]
• start branch model_summaries_91696941
• built branch model_summaries_91696941 [0.003 seconds]
• built pattern model_summaries
• end pipeline [0.151 seconds]And this time, when we load the model_summaries, we can
tell which model corresponds to which row (you may need to scroll to the
right to see it).
R
tar_read(model_summaries)
OUTPUT
# A tibble: 3 × 13
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual nobs model_name
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int> <chr>
1 0.0552 0.0525 1.92 19.9 1.12e- 5 1 -708. 1422. 1433. 1256. 340 342 combined_model
2 0.769 0.767 0.953 375. 3.65e-107 3 -467. 944. 963. 307. 338 342 species_model
3 0.770 0.766 0.955 225. 8.52e-105 5 -466. 947. 974. 306. 336 342 interaction_modelNext we will add one more target, a prediction of bill depth based on
each model. These will be needed for plotting the models in the report.
Such a prediction can be obtained with the augment()
function of the broom package.
R
tar_load(models)
augment(models[[1]])
OUTPUT
# A tibble: 342 × 8
bill_depth_mm bill_length_mm .fitted .resid .hat .sigma .cooksd .std.resid
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 18.7 39.1 17.6 1.14 0.00521 1.92 0.000924 0.594
2 17.4 39.5 17.5 -0.127 0.00485 1.93 0.0000107 -0.0663
3 18 40.3 17.5 0.541 0.00421 1.92 0.000168 0.282
4 19.3 36.7 17.8 1.53 0.00806 1.92 0.00261 0.802
5 20.6 39.3 17.5 3.06 0.00503 1.92 0.00641 1.59
6 17.8 38.9 17.6 0.222 0.00541 1.93 0.0000364 0.116
7 19.6 39.2 17.6 2.05 0.00512 1.92 0.00293 1.07
8 18.1 34.1 18.0 0.114 0.0124 1.93 0.0000223 0.0595
9 20.2 42 17.3 2.89 0.00329 1.92 0.00373 1.50
10 17.1 37.8 17.7 -0.572 0.00661 1.92 0.000296 -0.298
# ℹ 332 more rowsDefine the new function as augment_with_mod_name(). It
is the same as glance_with_mod_name(), but use
augment() instead of glance():
R
augment_with_mod_name <- function(model_in_list) {
model_name <- names(model_in_list)
model <- model_in_list[[1]]
augment(model) |>
mutate(model_name = model_name)
}
Add the step to the workflow:
R
source("R/functions.R")
source("R/packages.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance_with_mod_name(models),
pattern = map(models)
),
# Get model predictions
tar_target(
model_predictions,
augment_with_mod_name(models),
pattern = map(models)
)
)
Some other ways of applying branching patterns include:
- crossing: one branch per combination of elements
(
cross()function) - slicing: one branch for each of a manually selected set of elements
(
slice()function) - sampling: one branch for each of a randomly selected set of elements
(
sample()function)
You can find out
more about different branching patterns in the targets
manual.
Content from Parallel Processing
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How can we build targets in parallel?
Objectives
- Be able to build targets in parallel
Once a pipeline starts to include many targets, you may want to think about parallel processing. This takes advantage of multiple processors in your computer to build multiple targets at the same time.
targets includes support for high-performance computing,
cloud computing, and various parallel backends. Here, we assume you are
running this analysis on a laptop and so will use a relatively simple
backend. If you are interested in high-performance computing, see the
targets manual.
Install R packages for parallel computing
For this demo, we will use the new crew
backend.
Set up workflow
To enable parallel processing with crew you only need to
load the crew package, then tell targets to
use it using tar_option_set. Specifically, the following
lines enable crew, and tells it to use 2 parallel workers. You can
increase this number on more powerful machines:
R
library(crew)
tar_option_set(
controller = crew_controller_local(workers = 2)
)
Make these changes to the penguins analysis. It should now look like this:
R
source("R/functions.R")
source("R/packages.R")
# Set up parallelization
library(crew)
tar_option_set(
controller = crew_controller_local(workers = 2)
)
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance_with_mod_name(models),
pattern = map(models)
),
# Get model predictions
tar_target(
model_predictions,
augment_with_mod_name(models),
pattern = map(models)
)
)
There is still one more thing we need to modify only for the purposes of this demo: if we ran the analysis in parallel now, you wouldn’t notice any difference in compute time because the functions are so fast.
So let’s make “slow” versions of glance_with_mod_name()
and augment_with_mod_name() using the
Sys.sleep() function, which just tells the computer to wait
some number of seconds. This will simulate a long-running computation
and enable us to see the difference between running sequentially and in
parallel.
Add these functions to functions.R (you can copy-paste
the original ones, then modify them):
R
glance_with_mod_name_slow <- function(model_in_list) {
Sys.sleep(4)
model_name <- names(model_in_list)
model <- model_in_list[[1]]
broom::glance(model) |>
mutate(model_name = model_name)
}
augment_with_mod_name_slow <- function(model_in_list) {
Sys.sleep(4)
model_name <- names(model_in_list)
model <- model_in_list[[1]]
broom::augment(model) |>
mutate(model_name = model_name)
}
Then, change the plan to use the “slow” version of the functions:
R
source("R/functions.R")
source("R/packages.R")
# Set up parallelization
library(crew)
tar_option_set(
controller = crew_controller_local(workers = 2)
)
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance_with_mod_name_slow(models),
pattern = map(models)
),
# Get model predictions
tar_target(
model_predictions,
augment_with_mod_name_slow(models),
pattern = map(models)
)
)
Finally, run the pipeline with tar_make() as normal.
OUTPUT
✔ skip target penguins_data_raw_file
✔ skip target penguins_data_raw
✔ skip target penguins_data
✔ skip target models
• start branch model_predictions_5ad4cec5
• start branch model_predictions_c73912d5
• start branch model_predictions_91696941
• start branch model_summaries_5ad4cec5
• start branch model_summaries_c73912d5
• start branch model_summaries_91696941
• built branch model_predictions_5ad4cec5 [4.884 seconds]
• built branch model_predictions_c73912d5 [4.896 seconds]
• built branch model_predictions_91696941 [4.006 seconds]
• built pattern model_predictions
• built branch model_summaries_5ad4cec5 [4.011 seconds]
• built branch model_summaries_c73912d5 [4.011 seconds]
• built branch model_summaries_91696941 [4.011 seconds]
• built pattern model_summaries
• end pipeline [15.153 seconds]Notice that although the time required to build each individual target is about 4 seconds, the total time to run the entire workflow is less than the sum of the individual target times! That is proof that processes are running in parallel and saving you time.
The unique and powerful thing about targets is that we did not need to change our custom function to run it in parallel. We only adjusted the workflow. This means it is relatively easy to refactor (modify) a workflow for running sequentially locally or running in parallel in a high-performance context.
Now that we have demonstrated how this works, you can change your analysis plan back to the original versions of the functions you wrote.
Content from Reproducible Reports with Quarto
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- How can we create reproducible reports?
Objectives
- Be able to generate a report using
targets
Copy-paste vs. dynamic documents
Typically, you will want to communicate the results of a data analysis to a broader audience.
You may have done this before by copying and pasting statistics, plots, and other results into a text document or presentation. This may be fine if you only ever do the analysis once. But that is rarely the case—it is much more likely that you will tweak parts of the analysis or add new data and re-run your pipeline. With the copy-paste method, you’d have to remember what results changed and manually make sure everything is up-to-date. This is a perilous exercise!
Fortunately, targets provides functions for keeping a
document in sync with pipeline results, so you can avoid such pitfalls.
The main tool we will use to generate documents is
Quarto. Quarto can be used separately from
targets (and is a large topic on its own), but it also
happens to be an excellent way to dynamically generate reports with
targets.
Quarto allows you to insert the results of R code directly into your documents so that there is no danger of copy-and-paste mistakes. Furthermore, it can generate output from the same underlying script in multiple formats including PDF, HTML, and Microsoft Word.
Install Quarto
If you haven’t done so already, you will need to install Quarto, which is separate from R.
You will also need to install the quarto R package with
install.packages("quarto").
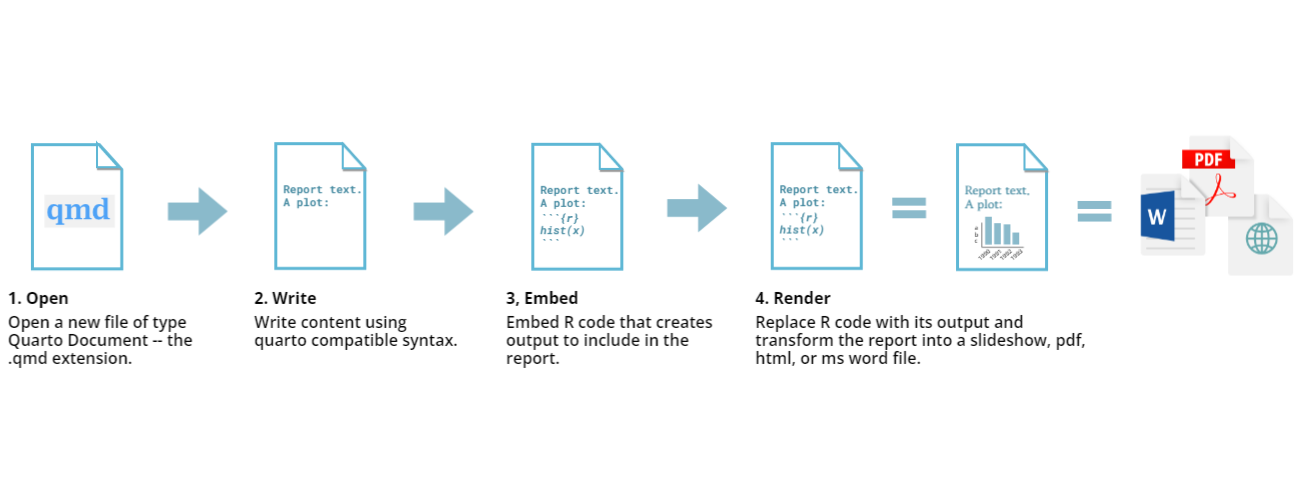
About Quarto files
.qmd or .Qmd is the extension for Quarto
files, and stands for “Quarto markdown”. Quarto files invert the normal
way of writing code and comments: in a typical R script, all text is
assumed to be R code, unless you preface it with a # to
show that it is a comment. In Quarto, all text is assumed to be prose,
and you use special notation to indicate which lines are R code to be
evaluated. Once the code is evaluated, the results get inserted into a
final, rendered document, which could be one of various formats.
We don’t have the time to go into the details of Quarto during this lesson, but recommend the “Introduction to Reproducible Publications with RStudio” incubator (in-development) lesson for more on this topic.
Recommended workflow
Dynamic documents like Quarto (or Rmarkdown, the predecessor to
Quarto) can actually be used to manage data analysis pipelines. But that
is not recommended because it doesn’t scale well and lacks the
sophisticated dependency tracking offered by targets.
Our suggested approach is to conduct the vast majority of data
analysis (in other words, the “heavy lifting”) in the
targets pipeline, then use the Quarto document to
summarize and plot the results.
Report on bill size in penguins
Continuing our penguin bill size analysis, let’s write a report evaluating each model.
To save time, the report is already available at https://github.com/joelnitta/penguins-targets.
Copy the raw
code from here and save it as a new file
penguin_report.qmd in your project folder (you may also be
able to right click in your browser and select “Save As”).
Then, add one more target to the pipeline using the
tar_quarto() function like this:
R
source("R/functions.R")
source("R/packages.R")
tar_plan(
# Load raw data
tar_file_read(
penguins_data_raw,
path_to_file("penguins_raw.csv"),
read_csv(!!.x, show_col_types = FALSE)
),
# Clean data
penguins_data = clean_penguin_data(penguins_data_raw),
# Build models
models = list(
combined_model = lm(
bill_depth_mm ~ bill_length_mm, data = penguins_data),
species_model = lm(
bill_depth_mm ~ bill_length_mm + species, data = penguins_data),
interaction_model = lm(
bill_depth_mm ~ bill_length_mm * species, data = penguins_data)
),
# Get model summaries
tar_target(
model_summaries,
glance_with_mod_name(models),
pattern = map(models)
),
# Get model predictions
tar_target(
model_predictions,
augment_with_mod_name(models),
pattern = map(models)
),
# Generate report
tar_quarto(
penguin_report,
path = "penguin_report.qmd",
quiet = FALSE,
packages = c("targets", "tidyverse")
)
)
The function to generate the report is tar_quarto(),
from the tarchetypes package.
As you can see, the “heavy” analysis of running the models is done in
the workflow, then there is a single call to render the report at the
end with tar_quarto().
How does targets know when to render the report?
It is not immediately apparent just from this how
targets knows to generate the report at the end of
the workflow (recall that build order is not determined by the
order of how targets are written in the workflow, but rather by their
dependencies). penguin_report does not appear to depend on
any of the other targets, since they do not show up in the
tar_quarto() call.
How does this work?
The answer lies inside the
penguin_report.qmd file. Let’s look at the start of the
file:
MARKDOWN
---
title: "Simpson's Paradox in Palmer Penguins"
format:
html:
toc: true
execute:
echo: false
---
```{r}
#| label: load
#| message: false
targets::tar_load(penguin_models_augmented)
targets::tar_load(penguin_models_summary)
library(tidyverse)
```
This is an example analysis of penguins on the Palmer Archipelago in Antarctica.The lines in between --- and --- at the
very beginning are called the “YAML header”, and contain directions
about how to render the document.
The R code to be executed is specified by the lines between
```{r} and ```. This is called a “code chunk”,
since it is a portion of code interspersed within prose text.
Take a closer look at the R code chunk. Notice the two calls to
targets::tar_load(). Do you remember what that function
does? It loads the targets built during the workflow.
Now things should make a bit more sense: targets knows
that the report depends on the targets built during the workflow,
penguin_models_augmented and
penguin_models_summary, because they are loaded in
the report with tar_load().
Generating dynamic content
The call to tar_load() at the start of
penguin_report.qmd is really the key to generating an
up-to-date report—once those are loaded from the workflow, we know that
they are in sync with the data, and can use them to produce “polished”
text and plots.
In the code chunk labeled
results-stats, statistics from the models like P-value and adjusted R squared are extracted, then inserted into the text with in-line code like`r mod_stats$combined$r.squared`.There are two figures, one for the combined model and one for the separate model (code chunks labeled
fig-combined-plotandfig-separate-plot, respectively). These are built using the points predicted from the model inpenguin_models_augmented.
You should also interactively run the code in
penguin_report.qmd to better understand what is going on,
starting with tar_load(). In fact, that is how this report
was written: the code was run in an interactive session, and saved to
the report as it was gradually tweaked to obtain the desired
results.
The best way to learn this approach to generating reports is to try it yourself.
So your final Challenge is to construct a targets
workflow using your own data and generate a report. Good luck!
